GaluxDesign is a pioneering platform for protein therapeutics design that uniquely integrates artificial intelligence with first principles.
We are developing next-generation AI for protein design by training it to understand the physical principles underlying protein folding and interactions, going beyond conventional data-driven interpolation and extrapolation.
The platform is modality-agnostic, meaning it can be applied to design a wide range of protein-based modalities including antibodies, VHHs, mini-proteins, cytokines and more. This broad compatibility accelerates innovation across diverse therapeutic areas.
By uniting physics-based AI and efficient experimental validation, GaluxDesign uncovers high-value leads that traditional methods often miss. This synergy with its tight coupling of AI and streamlined experimental validation, GaluxDesign evolves rapidly. The same integration also enables the efficient discovery of high-value molecules.
De novo protein design enables the creation of desirable proteins across a wide range of scaffolds, with the potential to open an entirely new landscape in protein therapeutics.
We are advancing GaluxDesign into a platform capable of zero-shot design of optimized molecules across diverse modalities, accounting for key properties such as binding affinity, stability, and immunogenicity.
In this case, GaluxDesign was used to generate antibodies targeting the atezolizumab epitope on PD-L1.The designed antibodies, with novel sequences, showed comparable binding affinity and early developability properties to the commercial reference antibody.
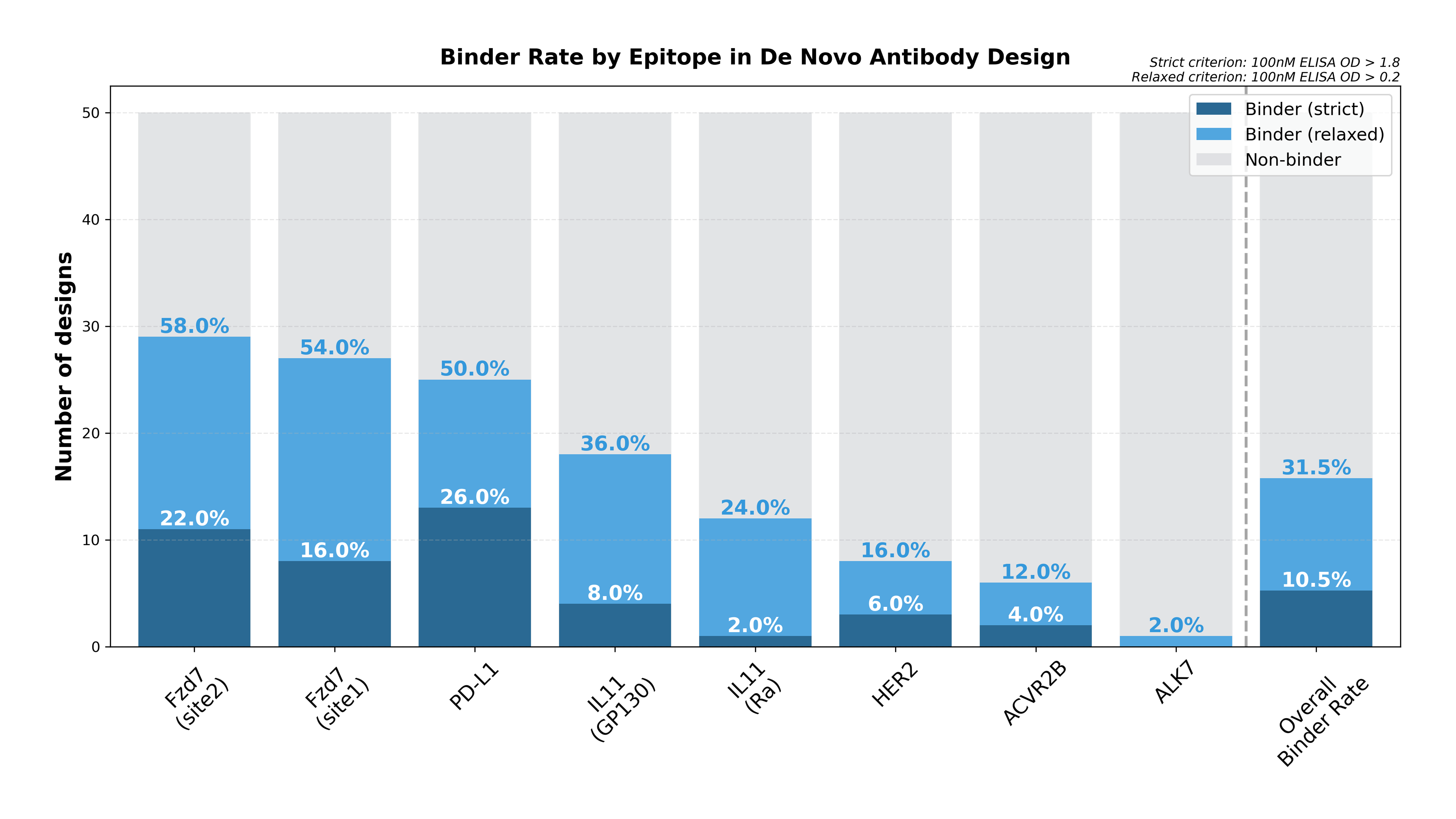
Our latest report on de novo antibody design is available on bioRxiv (link). In this article, we present successful cases of de novo antibody design against six different therapeutic targets, PD-L1, HER2, EGFR, FZD7, ACVR2A/2B, and ALK7. Notably, the FZD7 case (targeting a novel epitope) and the ALK7 case (designed without an experimentally determined structure) demonstrated the generalizability of GaluxDesign across diverse targets and design challenges.
To minimize immunogenicity risk and ensure favorable developability, all six antibodies were designed using clinically validated frameworks, with changes limited to the paratope regions.
In this case, a binding motif targeting the atezolizumab epitope on PD-L1 was created by engineering the terminus of the Fc region.
By designing a modality that lacks sequential and structural data entirely de novo, this example highlights the modality-agnostic nature of GaluxDesign.
Simultaneous optimization of multiple key properties enables more efficient and agile drug development. This approach can be applied to existing proteins to endow them with desirable properties such as binding affinity (including pH selectivity), specificity, and stability.
Insertion and deletion of amino acids can be freely incorporated, enabling broader exploration of optimal protein sequences and facilitating more flexible patent navigation.
In this case, IL-18 was engineered to achieve three key properties:
1. Elimination of binding to IL-18B
2. Enhanced binding to IL-18R
3. Improved thermal stability
With a few design-make-test cycles, the resulting IL-18 variants demonstrated the desirable properties. In a separate effort, we also explored immunocytokine designs with attenuated binding to IL-18R, systematically tuning affinity to identify the optimal therapeutic window.